Identifying network motifs for responses of chemical reaction networks

University of Illinois Urbana-Champaign,
Mathematics
Polly Yu
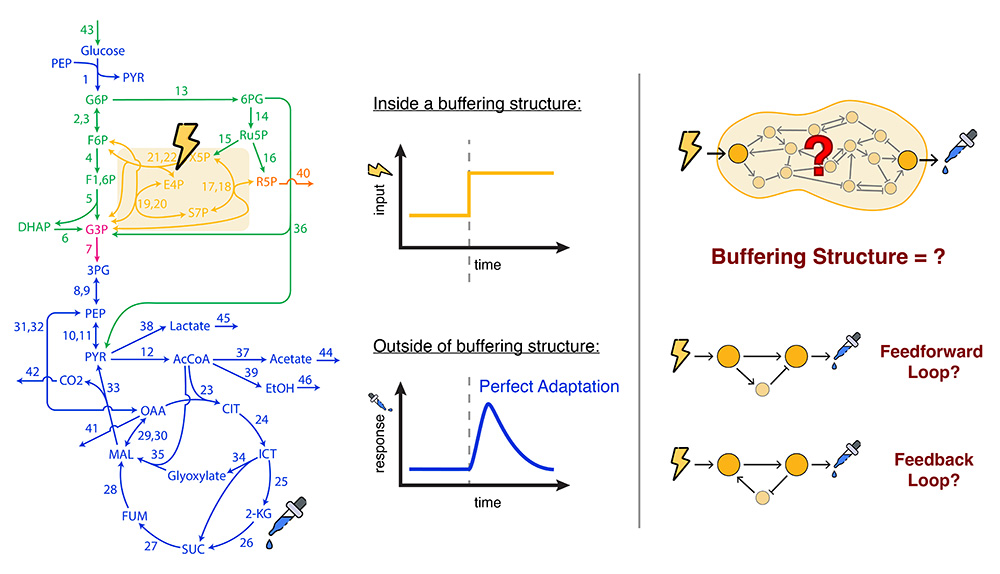
The biochemistry in living cells is known for its intricate dynamics, governed by overlapping reaction networks with many species. Despite the complexity, certain network structures appear repeatedly; these are known as network motifs, examples of which include feedback loops and feedforward loops. Links between such motifs and dynamics are evidence for the rich interplay between network structure and system behavior, which could be pivotal in understanding biological regulations.
Structural Sensitivity Analysis (SSA), as developed by the Mochizuki group, has successfully identified subnetworks (“buffering structures”) in biochemical reaction networks that prevent any perturbation within them from influencing the rest of the network. These subnetworks can be identified with subgraphs of the “species-reaction graph” of a reaction network, whose cycles and paths are intimately related to feedback and feedforward loops. We plan to find the connection between the subnetworks identified by SSA and the network motifs commonly referenced in synthetic and systems biology literature. This connection will serve as a dictionary between SSA, species-reaction graph, and the network motifs popular in biology.