RESEARCH
Theoretical studies for biological systems
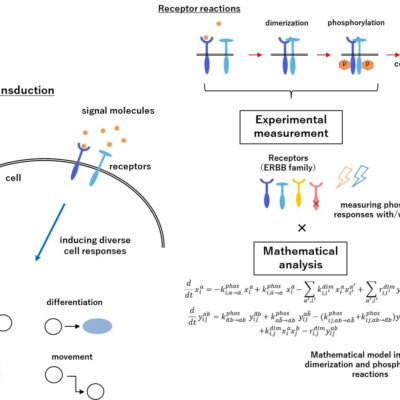
The progress of modern biology revealed that biological phenomena are governed by complex network systems including many molecules, cells or organs. For the aim of understanding the functions of complex systems, we adopt mathematical and computational methods. By theoretical approaches we decipher huge amounts of experimental information, and to give integrative understanding for the biological systems. Our final goal is to open a new bioscience which will progress by the repeats of the theoretical predictions and the experimental verifications. We are promoting multiple projects of collaborations with experimental biologists. One of our recent projects is studying dynamics of complex network systems in biology. We developed some theoretical frameworks to extract the important aspects of dynamics from network structure alone, without assuming other quantitative details. By combining our theory with experimental measuring and controling, we will determine mechanism of dynamical behaviours and understand the principles for the biological functions.
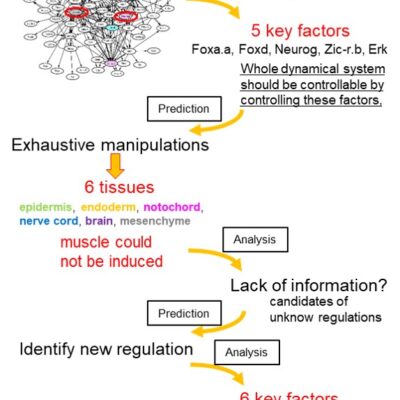
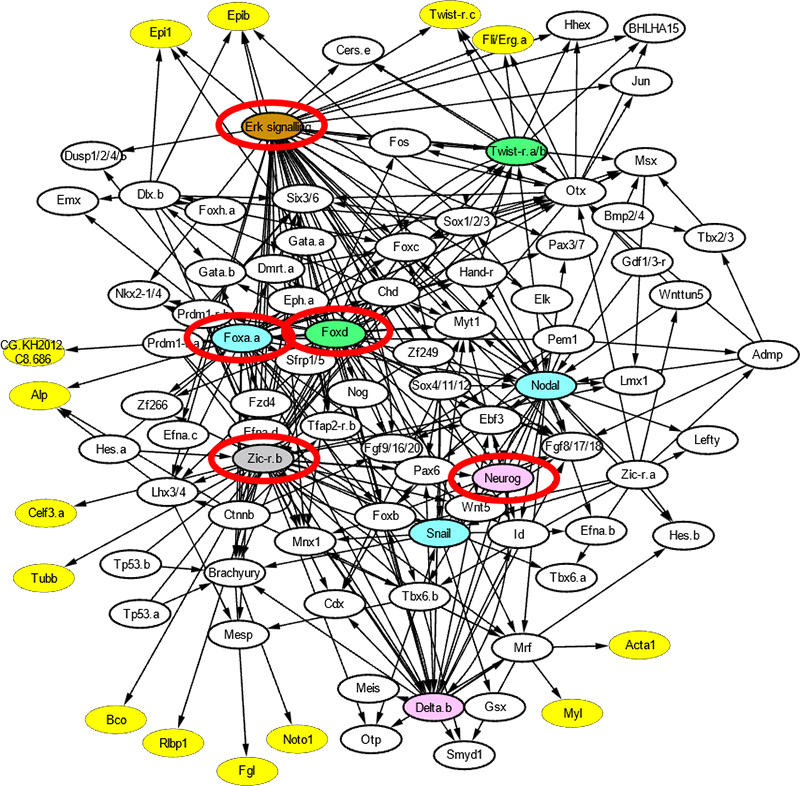
Gene regulatory network governing cell-fate specification of ascidian embryo
A small number of genes responsible for dynamical behaviors of the system was determined by the “Linkage Logic” theory. It is mathematically assured that the dynamical behaviors of the whole system are identifiable/controllable by observing/controlling behaviors of the red-marked genes.
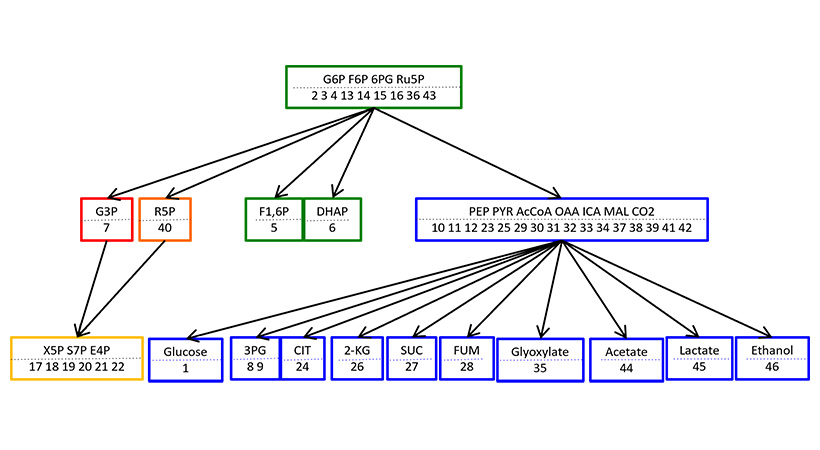
E. coli metabolic network
The vertices indicate metabolites, and edges indicate reactions. Colors summarize the response patterns shown in next Figure.
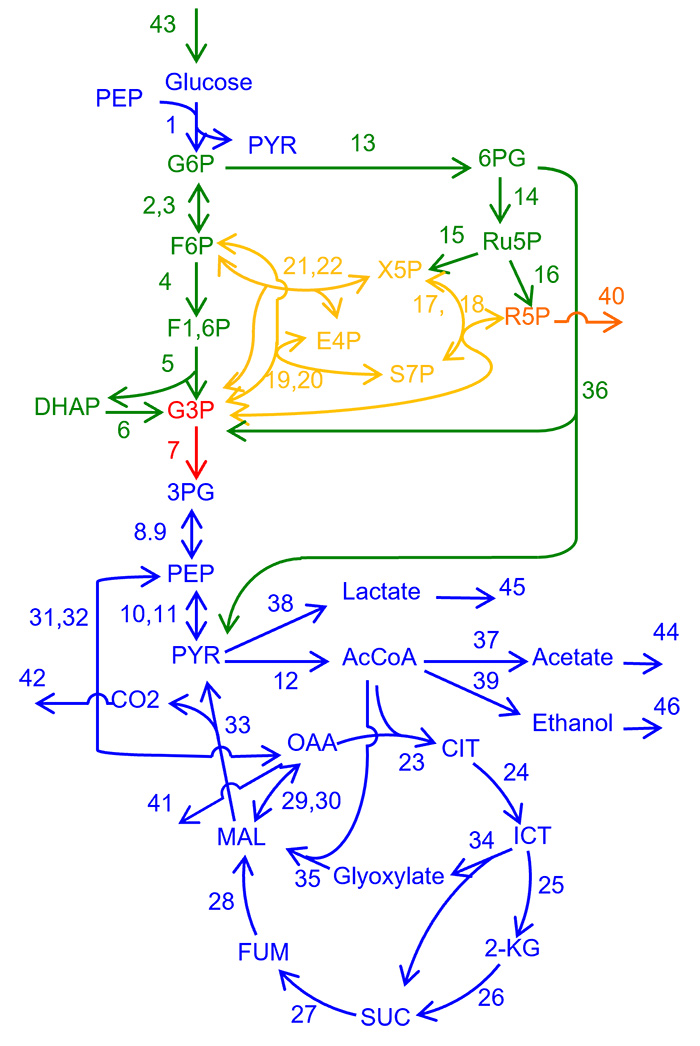
The “structural sensitivity analysis” clarify the modular and hierarchical structure in the control of metabolic system
Responses of chemical concentrations induced by changes in enzyme activities are localized in a finite range in the network, and show hierarchical inclusion relations. The ranges and hierarchy are determined from network structure, alone. The graph summarizes the response patterns. When activity of reaction enzyme shown in each square box is perturbed, the metabolites in the box plus those in the lower boxes show nonzero responses in their concentrations.